|
|
Mission Statement
|
High-throughput, next generation sequencing (NGS)
technology has transformed today's biological research
in many areas including genomics, transcriptomics and microbiomics. A variety of NGS sequencing
methods are available to satisfy a wide range of research requirements such as read length,
accuracy, speed, and cost. Accompanying the advent of NGS is the generation of large amount
of data. In this biological big data era, biologists are facing new challenges for data analysis
management and interpretation. Traditional bioinformatics software tools that run on a personal
computer often are crippled by the sheer size of the NGS sequencing data. High-performance
computational resource with multiple central processing units (CPUs), ample amount of memory,
and storage space with fast I/O (input/output, read/write) speed, are no longer luxurious but
necessary for doing research. Parallelly emerged in recent years in the field of computer and
information technology (IT) is the cloud computing technology - a computational model that
enables ubiquitous access to shared pools of computational resources such as networks, servers,
storage, applications and services. Cloud computing has gained a strong foothold in both
industry and science in the past decade and has risen to tackle the big data challenge.
The goal of Forsyth Bioinformatics Core is to help researchers meet this challenge and provide assistance
in big data analysis.
|
Who We Are
|
The Forsyth Bioinformatics Core was established
alongside the Human Oral Microbiome
Database (HOMD) since 2006. Led by
Dr. Tsute (George) Chen, a human oral microbiome data analysis expert, the core
has been developing bioinformatics tools to analyze the NGS data for research community
in the oral/dental and nasal study fields. The core houses an array of high-performance multi-CPU
computers with high memory that can analyze NGS data.
Two of our flagship NGS bioinformatics analysis pipelines are:
| 1. | Species-level classification and
profiling of 16S rRNA marker gene amplicon sequences
|
| | 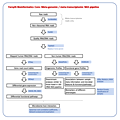 Click to view pipeline flowchart Click to view pipeline flowchart
|
|
| 2. | Organismic and functional profiling of meta-genomic and meta-transcriptomic NGS data
|
| |  Click to view pipeline flowchart Click to view pipeline flowchart
|
|
The key feature of our pipelines is the use of the HOMD curated
16S rRNA genes
and whole genome
reference sequences. Thus the results are tailored to, but not limited to, oral and nasal microbiological and clinical research fields.
Please follow this link to the publications involving these pipelines.
Check out some of the results using these two pipelines here.
We also provide consultation and analytical services in oral microbiomics, genomics, taxonomy, phylogenetics and all
next generation sequence (NGS) data analysis. The services are provided to both internal and external researchers
including the Harvard CATALYST community, for funded projects and
funding applications requiring preliminary bioinfromatics analysis or method writing.
|
| |
| |
Core Services
|
|
Next generation sequencing (NGS) data analysis
|
Microbiome profiling from 16S rRNA gene NGS data
|
|
Genomic sequencing raw reads assembly and annotation
|
RNAseq read mapping, transcriptome profiling and gene expression analysis
|
|
Metagenomic NGS sequence analysis
|
Metatranscriptomic NGS sequence Aanalysis
|
|
Grant writing and preliminary data analysis
|
Consultation on bioinformatics data analysis
|
|
| |
Service Inquiry and Request
|
|
We charge the service by hourly rate, by project, or by percent effort for a funded grant.
|
|
To request or inquire our services, please contact core director Dr. Tsute (George) Chen (Email: tchen@forsyth.org).
|
Forsyth Bioinformatics Core Publicatons
|
| A list of publications that the core coauthored is available here
|
|
| |
| |
Databases and Research Projects Supported by the Forsyth Bioinformatics Core
|
|
|
|
| |
| |
Software Tools Used by the Core
|
|
|
|
| |
| |
| |
| |
| |
| Copyright 2020 Forsyth Institute |
| Page last updated: December 10, 2020 21:13:04 | | |
|
|