|
|
FOMC Service Report
16S rRNA Gene V1V3 Amplicon Sequencing
Version V1.42
|
| The Forsyth Institute, Cambridge, MA, USA |
| November 17, 2022 |
|
Project ID: FOMC8810 |
|
I. Project Summary
|
|
Project FOMC8810 services include NGS sequencing of the V1V3 region of the 16S rRNA gene amplicons from the samples. First and foremost, please
download this report, as well as the sequence raw data from the download links provided below.
These links will expire after 60 days. We cannot guarantee the availability of your data after 60 days.
Bioinformatics analysis service was not requested, however we still provide the sequence data quality trimming, noise-filtering, pair merging, as well as chimera filtering for the sequences, using the
DADA2 denoising algorithm and pipeline. The denoised, merged and chimera-free ASV (amplicon sequence variants) sequences allow you to perform
downstream analyses such as taxonomy assignment, diversity analysis and differential abundance analysis. If you need us help with these downstream bioinformatics analysis please contact us.
|
|
| |
II. Workflow Checklist
|
| ☑ | 1. | Sample Received |
| ☑ | 2. | Sample Quality Evaluated |
| ☑ | 3. | Sample Prepared for Sequencing |
| ☑ | 4. | Next-Gen Sequencing |
| ☑ | 5. | Sequence Quality Check |
| ☑ | 6. | Absolute Abundance |
| ☑ | 7. | Report and Raw Sequence Data Available for Download |
| ☑ | 8. | Bioinformatics Analysis - Reads Processing (DADA2 Quality Trimming, Denoising, Paired Reads Merging) |
| ☐ | 9. | Bioinformatics Analysis - Reads Taxonomy Assignment (service not requested) |
| ☐ | 10. | Bioinformatics Analysis - Alpha Diversity Analysis (service not requested) |
| ☐ | 11. | Bioinformatics Analysis - Beta Diversity Analysis (service not requested) |
| ☐ | 12. | Bioinformatics Analysis - Differential Abundance Analysis (service not requested) |
| ☐ | 13. | Bioinformatics Analysis - Heatmap Profile (service not requested) |
| ☐ | 14. | Bioinformatics Analysis - Network Association (service not requested) |
|
|
| |
III. NGS Sequencing |
The samples were processed and analyzed with the ZymoBIOMICS® Service: Targeted
Metagenomic Sequencing (Zymo Research, Irvine, CA).
DNA Extraction: If DNA extraction was performed, one of three different DNA
extraction kits was used depending on the sample type and sample volume and were
used according to the manufacturer’s instructions, unless otherwise stated. The kit used
in this project is marked below:
| ☐ | ZymoBIOMICS® DNA Miniprep Kit (Zymo Research, Irvine, CA) |
| ☐ | ZymoBIOMICS® DNA Microprep Kit (Zymo Research, Irvine, CA) |
| ☐ | ZymoBIOMICS®-96 MagBead DNA Kit (Zymo Research, Irvine, CA) |
| ☑ | N/A (DNA Extraction Not Performed) |
| Elution Volume: 50µL |
| Additional Notes: NA |
Targeted Library Preparation: The DNA samples were prepared for targeted
sequencing with the Quick-16S™ NGS Library Prep Kit (Zymo Research, Irvine, CA).
These primers were custom designed by Zymo Research to provide the best coverage
of the 16S gene while maintaining high sensitivity. The primer sets used in this project
are marked below:
| ☐ | Quick-16S™ Primer Set V1-V2 (Zymo Research, Irvine, CA) |
| ☑ | Quick-16S™ Primer Set V1-V3 (Zymo Research, Irvine, CA) |
| ☐ | Quick-16S™ Primer Set V3-V4 (Zymo Research, Irvine, CA) |
| ☐ | Quick-16S™ Primer Set V4 (Zymo Research, Irvine, CA) |
| ☐ | Quick-16S™ Primer Set V6-V8 (Zymo Research, Irvine, CA) |
| ☐ | Other: NA |
| Additional Notes: NA |
The sequencing library was prepared using an innovative library preparation process in
which PCR reactions were performed in real-time PCR machines to control cycles and
therefore limit PCR chimera formation. The final PCR products were quantified with
qPCR fluorescence readings and pooled together based on equal molarity. The final
pooled library was cleaned up with the Select-a-Size DNA Clean & Concentrator™
(Zymo Research, Irvine, CA), then quantified with TapeStation® (Agilent Technologies,
Santa Clara, CA) and Qubit® (Thermo Fisher Scientific, Waltham, WA).
Control Samples: The ZymoBIOMICS® Microbial Community Standard (Zymo
Research, Irvine, CA) was used as a positive control for each DNA extraction, if
performed. The ZymoBIOMICS® Microbial Community DNA Standard (Zymo Research,
Irvine, CA) was used as a positive control for each targeted library preparation.
Negative controls (i.e. blank extraction control, blank library preparation control) were
included to assess the level of bioburden carried by the wet-lab process.
Sequencing: The final library was sequenced on Illumina® MiSeq™ with a V3 reagent kit
(600 cycles). The sequencing was performed with 10% PhiX spike-in.
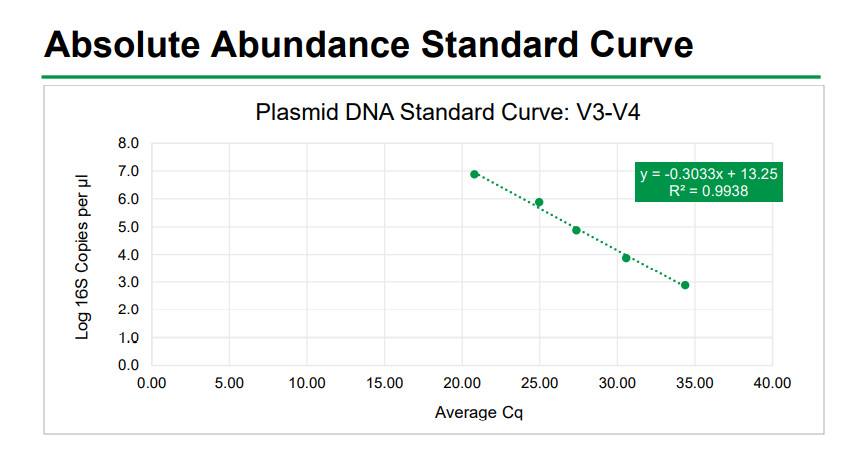
Absolute Abundance Quantification*: A quantitative real-time PCR was set up with a
standard curve. The standard curve was made with plasmid DNA containing one copy
of the 16S gene and one copy of the fungal ITS2 region prepared in 10-fold serial
dilutions. The primers used were the same as those used in Targeted Library
Preparation. The equation generated by the plasmid DNA standard curve was used to
calculate the number of gene copies in the reaction for each sample. The PCR input
volume (2 µl) was used to calculate the number of gene copies per microliter in each
DNA sample.
The number of genome copies per microliter DNA sample was calculated by dividing
the gene copy number by an assumed number of gene copies per genome. The value
used for 16S copies per genome is 4. The value used for ITS copies per genome is 200.
The amount of DNA per microliter DNA sample was calculated using an assumed
genome size of 4.64 x 106 bp, the genome size of Escherichia coli, for 16S samples, or
an assumed genome size of 1.20 x 107 bp, the genome size of Saccharomyces
cerevisiae, for ITS samples. This calculation is shown below:
Calculated Total DNA = Calculated Total Genome Copies × Assumed Genome Size (4.64 × 106 bp) ×
Average Molecular Weight of a DNA bp (660 g/mole/bp) ÷ Avogadro’s Number (6.022 x 1023/mole)
* Absolute Abundance Quantification is only available for 16S and ITS analyses.
The absolute abundance standard curve data can be viewed in Excel here:
The absolute abundance standard curve is shown below:
Absolute Abundance Standard Curve
|
|
| |
IV. Complete Report Download |
|
The complete report of your project, including all links in this report, can be downloaded by clicking the link provided below. The downloaded file is a compressed ZIP file and once unzipped, open the file “REPORT.html” (may only shown as "REPORT" in your computer) by double clicking it. Your default web browser will open it and you will see the exact content of this report.
Please download and save the file to your computer storage device. The download link will expire after 60 days upon your receiving of this report.
Complete report download link:
To view the report, please follow the following steps:
| 1. | Download the .zip file from the report link above. |
| 2. | Extract all the contents of the downloaded .zip file to your desktop. |
| 3. | Open the extracted folder and find the "REPORT.html" (may shown as only "REPORT"). |
| 4. | Open (double-clicking) the REPORT.html file. Your default browser will open the top age of the complete report. Within the
report, there are links to view all the analyses performed for the project. |
|
|
| |
V. Raw Sequence Data Download |
|
The raw NGS sequence data is available for download with the link provided below. The data is a compressed ZIP file and can be unzipped to individual sequence files.
Since this is a pair-end sequencing, each of your samples is represented by two sequence files, one for READ 1,
with the file extension “*_R1.fastq.gz”, another READ 2, with the file extension “*_R1.fastq.gz”.
The files are in FASTQ format and are compressed. FASTQ format is a text-based data format for storing both a biological sequence
and its corresponding quality scores. Most sequence analysis software will be able to open them.
The Sample IDs associated with the R1 and R2 fastq files are listed in the table below:
| Sample ID | Original Sample ID | Read 1 File Name | Read 2 File Name |
|---|
| F8810.S10 | original sample ID here | zr8810_10V3V4_R1.fastq.gz | zr8810_10V3V4_R2.fastq.gz |
| F8810.S11 | original sample ID here | zr8810_11V3V4_R1.fastq.gz | zr8810_11V3V4_R2.fastq.gz |
| F8810.S12 | original sample ID here | zr8810_12V3V4_R1.fastq.gz | zr8810_12V3V4_R2.fastq.gz |
| F8810.S13 | original sample ID here | zr8810_13V3V4_R1.fastq.gz | zr8810_13V3V4_R2.fastq.gz |
| F8810.S14 | original sample ID here | zr8810_14V3V4_R1.fastq.gz | zr8810_14V3V4_R2.fastq.gz |
| F8810.S15 | original sample ID here | zr8810_15V3V4_R1.fastq.gz | zr8810_15V3V4_R2.fastq.gz |
| F8810.S16 | original sample ID here | zr8810_16V3V4_R1.fastq.gz | zr8810_16V3V4_R2.fastq.gz |
| F8810.S17 | original sample ID here | zr8810_17V3V4_R1.fastq.gz | zr8810_17V3V4_R2.fastq.gz |
| F8810.S18 | original sample ID here | zr8810_18V3V4_R1.fastq.gz | zr8810_18V3V4_R2.fastq.gz |
| F8810.S19 | original sample ID here | zr8810_19V3V4_R1.fastq.gz | zr8810_19V3V4_R2.fastq.gz |
| F8810.S01 | original sample ID here | zr8810_1V3V4_R1.fastq.gz | zr8810_1V3V4_R2.fastq.gz |
| F8810.S20 | original sample ID here | zr8810_20V3V4_R1.fastq.gz | zr8810_20V3V4_R2.fastq.gz |
| F8810.S21 | original sample ID here | zr8810_21V3V4_R1.fastq.gz | zr8810_21V3V4_R2.fastq.gz |
| F8810.S22 | original sample ID here | zr8810_22V3V4_R1.fastq.gz | zr8810_22V3V4_R2.fastq.gz |
| F8810.S23 | original sample ID here | zr8810_23V3V4_R1.fastq.gz | zr8810_23V3V4_R2.fastq.gz |
| F8810.S24 | original sample ID here | zr8810_24V3V4_R1.fastq.gz | zr8810_24V3V4_R2.fastq.gz |
| F8810.S02 | original sample ID here | zr8810_2V3V4_R1.fastq.gz | zr8810_2V3V4_R2.fastq.gz |
| F8810.S03 | original sample ID here | zr8810_3V3V4_R1.fastq.gz | zr8810_3V3V4_R2.fastq.gz |
| F8810.S04 | original sample ID here | zr8810_4V3V4_R1.fastq.gz | zr8810_4V3V4_R2.fastq.gz |
| F8810.S05 | original sample ID here | zr8810_5V3V4_R1.fastq.gz | zr8810_5V3V4_R2.fastq.gz |
| F8810.S06 | original sample ID here | zr8810_6V3V4_R1.fastq.gz | zr8810_6V3V4_R2.fastq.gz |
| F8810.S07 | original sample ID here | zr8810_7V3V4_R1.fastq.gz | zr8810_7V3V4_R2.fastq.gz |
| F8810.S08 | original sample ID here | zr8810_8V3V4_R1.fastq.gz | zr8810_8V3V4_R2.fastq.gz |
| F8810.S09 | original sample ID here | zr8810_9V3V4_R1.fastq.gz | zr8810_9V3V4_R2.fastq.gz |
Please download and save the file to your computer storage device. The download link will expire after 60 days upon your receiving of this report.
Raw sequence data download link:
|
|
| |
VI. Analysis - DADA2 Read Processing |
What is DADA2?
DADA2 is a software package that models and corrects Illumina-sequenced amplicon errors.
DADA2 infers sample sequences exactly, without coarse-graining into OTUs,
and resolves differences of as little as one nucleotide. DADA2 identified more real variants
and output fewer spurious sequences than other methods.
DADA2’s advantage is that it uses more of the data. The DADA2 error model incorporates quality information,
which is ignored by all other methods after filtering. The DADA2 error model incorporates quantitative abundances,
whereas most other methods use abundance ranks if they use abundance at all.
The DADA2 error model identifies the differences between sequences, eg. A->C,
whereas other methods merely count the mismatches. DADA2 can parameterize its error model from the data itself,
rather than relying on previous datasets that may or may not reflect the PCR and sequencing protocols used in your study.
DADA2 Publication:
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016 Jul;13(7):581-3. doi: 10.1038/nmeth.3869. Epub 2016 May 23. PMID: 27214047; PMCID: PMC4927377.
DADA2 Software Package is available as an R package at : https://benjjneb.github.io/dada2/index.html
Analysis Procedures:
DADA2 pipeline includes several tools for read quality control, including quality filtering, trimming, denoising, pair merging and chimera filtering. Below are the major processing steps of DADA2:
Step 1. Read trimming based on sequence quality
The quality of NGS Illumina sequences often decreases toward the end of the reads.
DADA2 allows to trim off the poor quality read ends in order to improve the error
model building and pair mergicing performance.
Step 2. Learn the Error Rates
The DADA2 algorithm makes use of a parametric error model (err) and every
amplicon dataset has a different set of error rates. The learnErrors method
learns this error model from the data, by alternating estimation of the error
rates and inference of sample composition until they converge on a jointly
consistent solution. As in many machine-learning problems, the algorithm must
begin with an initial guess, for which the maximum possible error rates in
this data are used (the error rates if only the most abundant sequence is
correct and all the rest are errors).
Step 3. Infer amplicon sequence variants (ASVs) based on the error model built in previous step. This step is also called sequence "denoising".
The outcome of this step is a list of ASVs that are the equivalent of oligonucleotides.
Step 4. Merge paired reads. If the sequencing products are read pairs, DADA2 will merge the R1 and R2 ASVs into single sequences.
Merging is performed by aligning the denoised forward reads with the reverse-complement of the corresponding
denoised reverse reads, and then constructing the merged “contig” sequences.
By default, merged sequences are only output if the forward and reverse reads overlap by
at least 12 bases, and are identical to each other in the overlap region (but these conditions can be changed via function arguments).
Step 5. Remove chimera.
The core dada method corrects substitution and indel errors, but chimeras remain. Fortunately, the accuracy of sequence variants
after denoising makes identifying chimeric ASVs simpler than when dealing with fuzzy OTUs.
Chimeric sequences are identified if they can be exactly reconstructed by
combining a left-segment and a right-segment from two more abundant “parent” sequences. The frequency of chimeric sequences varies substantially
from dataset to dataset, and depends on on factors including experimental procedures and sample complexity.
Results
1. Read Quality Plots NGS sequence analaysis starts with visualizing the quality of the sequencing. Below are the quality plots of the first
sample for the R1 and R2 reads separately. In gray-scale is a heat map of the frequency of each quality score at each base position. The mean
quality score at each position is shown by the green line, and the quartiles of the quality score distribution by the orange lines.
The forward reads are usually of better quality. It is a common practice to trim the last few nucleotides to avoid less well-controlled errors
that can arise there. The trimming affects the downstream steps including error model building, merging and chimera calling. FOMC uses an empirical
approach to test many combinations of different trim length in order to achieve best final amplicon sequence variants (ASVs), see the next
section “Optimal trim length for ASVs”.
Quality plots for all samples:
2. Optimal trim length for ASVs The final number of merged and chimera-filtered ASVs depends on the quality filtering (hence trimming) in the very beginning of the DADA2 pipeline.
In order to achieve highest number of ASVs, an empirical approach was used -
- Create a random subset of each sample consisting of 5,000 R1 and 5,000 R2 (to reduce computation time)
- Trim 10 bases at a time from the ends of both R1 and R2 up to 50 bases
- For each combination of trimmed length (e.g., 300x300, 300x290, 290x290 etc), the trimmed reads are
subject to the entire DADA2 pipeline for chimera-filtered merged ASVs
- The combination with highest percentage of the input reads becoming final ASVs is selected for the complete set of data
Below is the result of such operation, showing ASV percentages of total reads for all trimming combinations (1st Column = R1 lengths in bases; 1st Row = R2 lengths in bases):
|
| R1/R2 | 271 | 261 | 251 | 241 | 231 |
|---|
| 321 | 12.37% | 14.92% | 20.92% | 22.28% | 26.48% |
|---|
| 311 | 14.49% | 19.32% | 27.69% | 30.62% | 33.78% |
|---|
| 301 | 13.61% | 19.05% | 27.86% | 31.29% | 34.92% |
|---|
| 291 | 13.56% | 18.59% | 26.64% | 31.25% | 34.23% |
|---|
| 281 | 13.52% | 17.79% | 26.49% | 30.93% | 33.70% |
|---|
| 271 | 13.56% | 17.88% | 26.08% | 30.55% | 33.14% |
|---|
|
|
Based on the above result, the trim length combination of R1 = 301 bases and R2 = 231 bases (highlighted red above), was chosen for generating final ASVs for all sequences.
This combination generated highest number of merged non-chimeric ASVs and was used for downstream analyses, if requested.
3. Error plots from learning the error rates
After DADA2 building the error model for the set of data, it is always worthwhile, as a sanity check if nothing else, to visualize the estimated error rates.
The error rates for each possible transition (A→C, A→G, …) are shown below. Points are the observed error rates for each consensus quality score.
The black line shows the estimated error rates after convergence of the machine-learning algorithm.
The red line shows the error rates expected under the nominal definition of the Q-score.
The ideal result would be the estimated error rates (black line) are a good fit to the observed rates (points), and the error rates drop
with increased quality as expected.
Forward Read R1 Error Plot
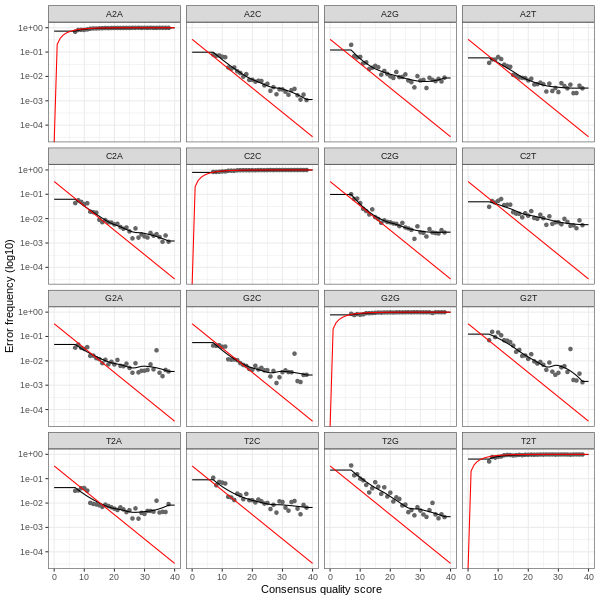 Reverse Read R2 Error Plot
Reverse Read R2 Error Plot
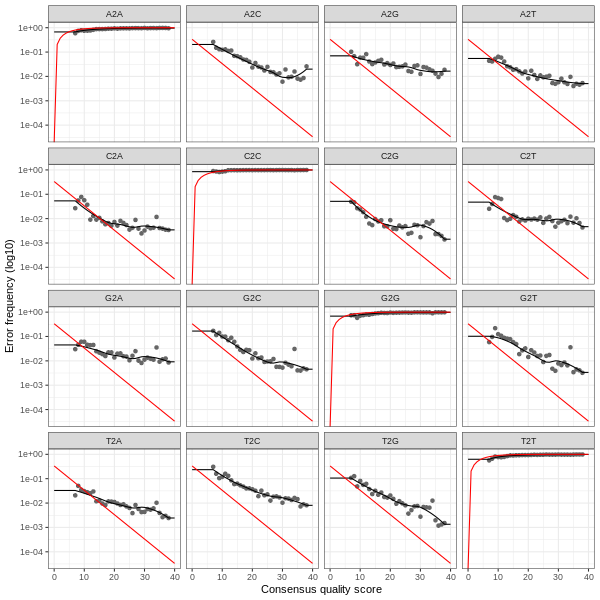
The PDF version of these plots are available here:
|
| |
|
4. DADA2 Result Summary The table below shows the summary of the DADA2 analysis,
tracking paired read counts of each samples for all the steps during DADA2 denoising process -
including end-trimming (filtered), denoising (denoisedF, denoisedF), pair merging (merged) and chimera removal (nonchim).
|
| Sample ID | F8810.S01 | F8810.S02 | F8810.S03 | F8810.S04 | F8810.S05 | F8810.S06 | F8810.S07 | F8810.S08 | F8810.S09 | F8810.S10 | F8810.S11 | F8810.S12 | F8810.S13 | F8810.S14 | F8810.S15 | F8810.S16 | F8810.S17 | F8810.S18 | F8810.S19 | F8810.S20 | F8810.S21 | F8810.S22 | F8810.S23 | F8810.S24 | Row Sum | Percentage |
|---|
| input | 70,590 | 63,446 | 68,819 | 63,670 | 59,326 | 73,510 | 68,395 | 78,728 | 71,134 | 40,072 | 70,970 | 70,706 | 74,170 | 65,198 | 54,688 | 67,931 | 76,491 | 69,085 | 69,302 | 49,680 | 56,821 | 64,341 | 75,305 | 71,296 | 1,593,674 | 100.00% |
|---|
| filtered | 70,588 | 63,446 | 68,818 | 63,670 | 59,325 | 73,510 | 68,394 | 78,727 | 71,133 | 40,071 | 70,970 | 70,706 | 74,170 | 65,197 | 54,687 | 67,931 | 76,491 | 69,084 | 69,302 | 49,680 | 56,819 | 64,341 | 75,304 | 71,294 | 1,593,658 | 100.00% |
|---|
| denoisedF | 64,786 | 57,835 | 64,917 | 58,742 | 53,261 | 67,209 | 62,752 | 76,988 | 64,937 | 37,318 | 65,030 | 65,451 | 66,846 | 59,057 | 49,380 | 63,528 | 71,226 | 62,011 | 66,339 | 48,262 | 52,380 | 58,035 | 69,586 | 64,425 | 1,470,301 | 92.26% |
|---|
| denoisedR | 67,113 | 59,510 | 65,616 | 59,325 | 55,815 | 69,291 | 62,285 | 76,698 | 67,990 | 37,997 | 61,553 | 67,782 | 68,207 | 58,119 | 50,091 | 62,361 | 72,402 | 64,881 | 66,624 | 47,297 | 54,301 | 56,771 | 71,307 | 67,754 | 1,491,090 | 93.56% |
|---|
| merged | 48,268 | 43,675 | 51,402 | 41,978 | 37,425 | 49,993 | 48,202 | 62,010 | 51,455 | 31,791 | 41,685 | 54,863 | 46,831 | 38,902 | 32,680 | 49,439 | 56,206 | 46,860 | 38,805 | 39,935 | 41,046 | 38,356 | 54,129 | 42,857 | 1,088,793 | 68.32% |
|---|
| nonchim | 25,431 | 26,094 | 13,943 | 19,276 | 23,406 | 30,159 | 23,491 | 6,836 | 23,490 | 10,260 | 24,778 | 26,154 | 25,430 | 24,407 | 21,194 | 15,505 | 19,503 | 30,190 | 13,816 | 5,946 | 17,579 | 23,447 | 23,396 | 25,301 | 499,032 | 31.31% |
|---|
|
This table can be downloaded as an Excel table below:
|
| |
|
5. DADA2 Amplicon Sequence Variants (ASVs). A total of 2998 unique merged and chimera-free ASV sequences were identified, and their corresponding
read counts for each sample are available in the "ASV Read Count Table" with rows for the ASV sequences and columns for sample. This read count table can be used for
microbial profile comparison among different samples and the sequences provided in the table can be used to taxonomy assignment.
|
| |
|
The table can be downloaded from this link:
|
| |
|
| |
VII. Analysis - Read Taxonomy Assignment |
| Service not requested |
| |
VIII. Analysis - Alpha Diversity |
| Service not requested |
| |
IX. Analysis - Beta Diversity |
| Service not requested |
| |
X. Analysis - Differential Abundance |
| Service not requested |
| |
XI. Analysis - Heatmap Profile |
| Service not requested |
| |
XII. Analysis - Network Association |
| Service not requested |
| |
|
| Copyright FOMC 2022
|
|
|
|

